University of Missouri St. Louis/ Biology Department/ Faculty/ Dr. Wendy M. Olivas/
Our Research
University of Missouri St. Louis/ Biology Department/ Faculty/ Dr. Wendy M. Olivas/
Our Research

within the 3' untranslated region (3' UTR) of an mRNA.
Several types of regulatory proteins have been identified that bind these
control elements, yet little is known about how the binding of these factors
leads to functional changes in the behavior of the mRNAs. We are studying
how members of the Puf family of proteins are able to bind and
differentially regulate the decay rate of several target mRNAs. Puf proteins
appear to have a conserved role in stem cell maintenance, cell development,
and differentiation, and are found across the eukaryotic kingdom. However,
information about how any of these Puf proteins influence the mRNA decay
machinery is only beginning to be uncovered.
The yeast Saccharomyces cerevisiae is a model organism for understanding how
Puf proteins modulate mRNA lifespans. S. cerevisiae contains six Puf
proteins, providing a unique system for studying differential Puf protein
target specificity and regulatory function. In addition, the general
pathways of mRNA decay in yeast are well described, and multiple lines of
evidence suggest that these pathways are conserved among eukaryotes. Our
analysis of the mechanisms by which the yeast Puf proteins recognize and
regulate the stability of their target mRNAs will greatly contribute to our
understanding of Puf-mediated control of gene expression in yeast and other
eukaryotes, and to the general principles of 3' UTR control of mRNA decay. A
fundamental issue in understanding mRNA-specific regulation by Puf proteins
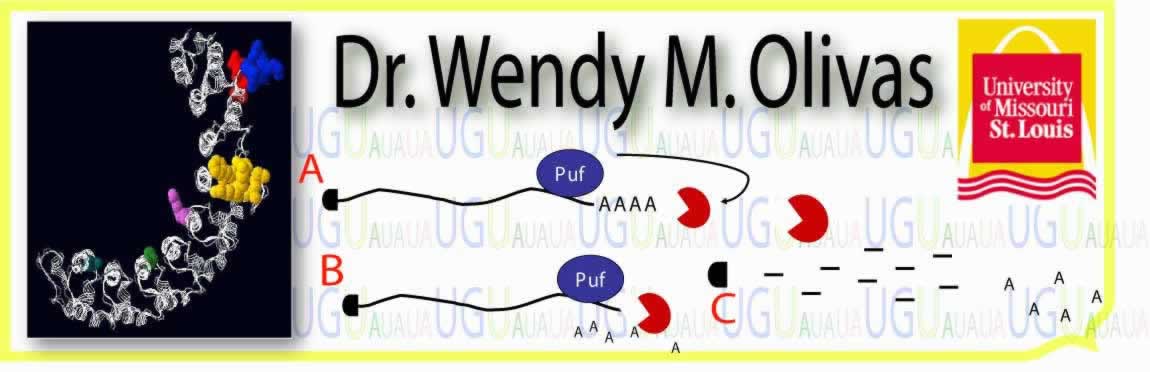
is the determination of the 3' UTR elements involved . We had earlier
identified Puf3p as the first transcript-specific regulator of mRNA
deadenylation and decay in yeast (Olivas and Parker, EMBO J. , 2000).
Therefore, we characterized the role of specific nucleotides within the 3'
UTR of the COX17 mRNA that are responsible and sufficient for binding the
Puf3 protein and promoting mRNA decay (Jackson et al., RNA , 2004). Our
analysis identified two distinct Puf3p binding sites containing a conserved
sequence element. The two sites show differential binding affinity to Puf3p
in vitro, with sequences flanking the conserved binding element influencing
affinity. However, each site is equally sufficient for partial stimulation
of COX17 mRNA decay in vivo, while full decay regulation requires both
sites. This indicates that multiple Puf3p signals can combine to increase
decay activity. No other sequences outside the 3' UTR are required for
Puf3p-mediated decay. In addition, we found the repeat domain of Puf3p to be
sufficient for both RNA binding and mRNA decay stimulation, supporting a
conserved role of the Puf repeat domain as an independent regulator of mRNA
metabolism.
Another important issue in understanding
mRNA-specific regulation by Puf proteins is determining how different Puf
proteins that are well conserved in their repeat domains target different
mRNAs for decay. Thus, we identified the Puf3p amino acid positions that are
involved in target-specific binding to the COX17 mRNA, as well as protein
regions responsible for signaling to the mRNA decay machinery (Houshmandi
and Olivas, RNA , 2005). Crystal structure analysis of a human Puf bound to
RNA had suggested a modular mode of binding, with three amino acid positions
within each of the eight repeats of the repeat domain contacting a single
nucleotide in the target RNA (Wang et al., Cell, 2002). However, by mutating
predicted RNA-binding positions of Puf3p to those found in Puf5p, we found
that a simple set of rules cannot reliably link specific amino acid
positions with target specificity. Moreover, we found that particular amino
acid positions played different roles within Puf3p and Puf5p for binding to
their respective target mRNAs. Finally, we identified an outer surface loop
that is dispensable for RNA binding, but required to promote mRNA decay.
The identification of additional genes required for Puf3p-mediated
stimulation of mRNA decay gets at the heart of the mechanism by which Puf
proteins influence the mRNA decay machinery. To this end, we identified
proteins that interact with Puf3p to mediate rapid mRNA decay. Specifically,
we identified several mRNA decay factors that interact with Puf3p, including
proteins involved in both deadenylation and mRNA decapping. All of these
interacting proteins were found to be essential for Puf3p-mediated COX17
decay, and all appear to bind Puf3p independent of RNA. We are currently
analyzing the mechanistic role of these protein interactions.
Another project in the lab has focused on the regulation of Puf protein
activity. In collaboration with Dr. Harmen Bussemaker's computational
biology lab at Columbia University , we made the first experimental report
of condition-specific regulation of Puf3p activity (Foat et al., PNAS ,
2005). Specifically, we found that Puf3p activity is inhibited by growth in
ethanol and rapamycin, though to different extents. We are currently
studying the mechanism by which Puf3p activity is altered under different
carbon source and other environmental conditions. We have also identified
several new targets of Puf3 protein regulation. Using this set of Puf3p
target mRNAs, we have established several different growth conditions that
alter Puf3p activity on these targets. Moreover, modulation of Puf3p
activity occurs very rapidly upon changes in growth conditions, providing
support for a post-translational control event.
Other studies have identified mRNA targets of the Puf4 protein, and
experiments are underway to analyze the condition-specific regulation of
Puf4p on these targets. Our results show that Puf4p activity is strongly
inhibited in several carbon source and stress conditions that only mildly
affect Puf3p activity. Current work aims to determine whether Puf4p and
Puf3p activities are altered by similar mechanisms. Unlike Puf3p, which
stimulates decay, our experiments indicate that Puf4p may act to stabilize
its target mRNAs. These results open a whole new avenue of research to
determine the mechanism by which Puf4p inhibits the mRNA decay machinery.
A complementary direction in the lab has concentrated on analyzing the
regulation of mRNA targets by the yeast Puf proteins, Puf1p, Puf2p, and
Puf5p. Several mRNAs whose levels and decay rates are affected by these Pufs
have been identified. In a novel finding, two mRNA targets were found that
are regulated in a combinatorial fashion by both Puf1p and Puf5p. Decay
regulation involves multiple binding elements in the 3' UTRs of the mRNAs,
and regulatory activity is condition-specific. Future work will involve the
analysis of the mechanism of mRNA regulation by these Puf proteins.
University of Missouri - St. Louis, One University Boulevard -
St. Louis, MO 63121-4400 USA 314-516-5000. - Contact Us- Maps
Last Update 1 December 2006.
Owner: Wendy M. Olivas
Our lab studies the regulation of messenger RNA decay in yeast, a model eukaryotic organism. Multiple processes within the cell mediate the proper control of gene expression. One key aspect of gene expression is the control of mRNA lifespans by regulation of mRNA decay rates. The decay rates of individual mRNAs in eukaryotic cells can vary by more than two orders of magnitude. Moreover, modulation of mRNA decay rates is an efficient method to rapidly alter gene expression in response to cellular changes. The control elements that mediate specific mRNA decay rates are often found