-
About Us
Keith J. Stine, Chair
Mission Statement
Degree Offerings
Department Photos
Outreach Activities
-
Undergraduate Studies
Undergraduate Degrees
Undergraduate Program Advising
Undergraduate Scholarships and Awards
Research for Undergraduates
Resources
-
Graduate Studies
Graduate Program Overview
Graduate Degrees
Application to Graduate Program
Graduate Awards
Graduate Program Contact Information
-
Faculty
Faculty Contact Information
Inorganic
Organic
Analytical/Physical
Biochemistry
Chemistry Faculty Awards
-
Staff
-
Seminar Programs
Archive
Graduate Student Seminars
Robert W. Murray Lecture
Distinguished Alumni Lecture
-
Departmental News
News Archive
-
Facilities
High Field NMR Facility
MIST Lab
X-Ray Diffraction Facility
-
Alumni Interests
UMSL Chemists
Important Dates
Distinguished Alumni
Alumni Lecturers
-
Contact Information
 |
Dr. Nichols received his B.S. degree from Lindenwood College and Ph.D. from Purdue University. Prior to joining UMSL in Fall 2004, he completed a postdoctoral fellowship at the Mayo Clinic in Jacksonville, FL. He was appointed Associate Chair in 2019. |
nicholsmic@umsl.edu |
Research Interests
Protein assembly or aggregation is widely recognized as a significant contributing factor to a number of neurodegenerative diseases including Alzheimer's disease (AD), Parkinson's disease, Huntington's disease, and others. Remarkably, the proteins or peptides implicated in these diseases, while possessing different amino acid sequences, all self-assemble to form similar fibrillar structures termed amyloid. One such peptide is amyloid-β (Aβ), a 40-42-residue peptide and the primary component of the senile plaques found in AD brains. The leading hypothesis in AD research maintains that accumulation of aggregated Aβ is the primary cause of the disease.
One research area in my laboratory involves mechanistic studies of Aβ aggregation. Objectives include isolation and characterization of aggregation intermediates and investigation of conditions that influence aggregation.These studies utilize an array of biophysical techniques to probe mechanistic and structural questions.
The other major research thrust in my laboratory addresses the question of how Aβ aggregates are detrimental to cells. One hypothesis is induction of a sustained inflammatory response causing the release of harmful cytokines such as tumor necrosis and factor-α interleukin-1-β. We are studying these effects in microglial and monocyte/macrophage cells with the goal of understanding the cause of the inflammatory response, how it relates to cell toxicity, and identification of novel ways to regulate these inflammatory pathways.
 |
 |
|
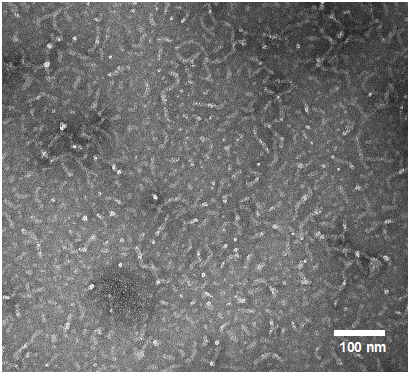
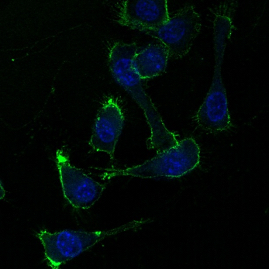
| (Left) Electron microscopy image (59k magnification) of isolated Aβ42 protofibrils. (Right) Binding of Aβ42 protofibrils (green) tp BV-2 microglia. Cell nuclei are shown in blue. | ||
Selected Publications
″A new class of monoclonal Aβ antibodies selectively targets and triggers deposition of Aβ protofibrils″ S. Grover, T. Pham, A. Jones, C. Sinobas-Pereira, M. Villoch Diaz Maurino, E. C. Garrad, N. J. Makoni, A. Parks, R. J. Domalewski, G. Riggio, H. An, K. Chen and M. R. Nichols. J. Neurochem. 2023 165, 860.
″Effect of mesoporous silica nanoparticles loaded with α-tomatine on HepG2 cancer cells studied in vitro″, B. Nepal, J. K. Bhattarai, K. Dharmi, M. R. Nichols and K. J. Stine, J. Drug. Deliv. Sci & Tech. 2023, 79, 104033.
″Development of β-sheet structure in Aβ aggregation intermediates diminishes exposed hydrophobic surface area and enhances proinflammatory activity,″ K. B. Dhami, S. Karki, A. Parks, C. G. Nichols, and M. R. Nichols, Biochim. Biophys. Acta, Proteins & Proteomics, 2022, 1870, 140817.
″Development of a Simple and Effective Lipid-A Antagonist Based on Computational Prediction," O. Slater, K. P. Dhami, G. Shrestha, M. Kontoyianni, M. R. Nichols and A. V. Demchenko, ACS Infect. Diseases, 2022, 8, 1171.
″Expression of NLRP3 inflammasome proteins in ExpiCHO-S mammalian cells reveals oligomerization properties that are highly sensitive to solution conditions,″ N. Y. Makoni, E. C. Garrad, A. Rezic and M. R. Nichols, Biotech Progress, 2021, 37, e3153.
″The intricate biophysical puzzle of caspase-1 activation,″ N. J. Makoni and M. R. Nichols, Arch. Biochem. Biophys. 2021, 699, 108753
″Inhibition of matrix metalloproteinase-9-secretion by dimethyl sulphoxide and cyclic adenosine monophosphate in human monocytes,″ D. R Denner, M. LD Udan-Johns and M. R Nichols, World J Biol Chem, 2021 12, 1
″Human and mouse single-nucleus transcriptomics reveal TREM2-dependent and TREM2-independent cellular responses in Alzheimer's disease,″ Y. Zhou, W. M. Song, P. S. Andhey, A. Swain, T. Levy, K. R. Miller, P. L. Poliani, M. Cominelli, S. Grover, S Gilfillan, M. Cella, T. K. Ulland, K. Zaitsev, A. Miyashita, T. Ikeuchi, M. Sainouchi, A. Kakita, D. A. Bennett, J. A. Schneider, M. R.Nichols, S. A. Beausoleil, J. D. Ulrich, D. M. Holtzman, M. N. Artyomov and M. Colonna, Nature Medicine 2020, 26, 131.
″Disentangling aggregation-prone proteins: a new method for isolating α-synuclein species, An Editorial Highlight for "A simple, versatile and robust centrifugation-based filtration protocol for the isolation and quantification of α-synuclein monomers, oligomers and fibrils: Towards improving experimental reproducibility in α-synuclein research," M. R. Nichols, J. Neurochem. 2020, 153, 7.
″Inflammatory mechanisms in neurodegeneration,″ M. R. Nichols, M-K. St-Pierre, A-C. Wendeln, N. J. Makoni, L. K. Gouwens, E. C. Garrad, M. Sohrabi, J. J. Neher, M-E Tremblay and C. K. Combs, J. Neurochem. 2019, 49, 562.
″Aβ42 Protofibrils Interact with and Are Trafficked through Microglial-Derived Microvesicles, ″ L. K. Gouwens, M. S. Ismail, V. A. Rogers, N. T. Zeller, E. C. Garrad, F. T. Amtashar, N. J. Makoni, D. C. Osborn and M. R. Nichols, ACS Chemical Neuroscience, 2018, 9, 1416
″The conformational epitope for a new Aβ42 protofibril-selective antibody partially overlaps with the peptide N-terminal region,″ B. A. Colvin, V. A. Rogers, J. A. Kulas, E. A. Ridgway, F. S. Amtashar, C. K. Combs and M. R. Nichols, J. Neurochem. 2017, 143, 736
″Amyloid-β42 protofibrils are internalized by microglia more extensively than monomers,″ L. K. Gouwens, N. J. Makoni, V. A. Rogers and M. R. Nichols, Brain Res., 2016, 1648, Part_A, 485.
″APP regulates microglial phenotype in a mouse model of Alzheimer's disease,″ G. D. Manocha, A. M. Floden, K. Rausch, J. A. Kulas, B. A. McGregor, L. Rojanathammanee, K. R. Puig, K. L. Puig, S. Karki, J. E. Porter and C. K. Combs, M. R. Nichols, D. C. Darland, J. Neuroscience, 2016, 36, 8471
″Aβ40 has a subtle effect on Aβ42 protofibril formation, but to a lesser degree than Aβ42 concentration, in Aβ42/Aβ40 mixtures,″ S. E. Terrill-Usery, B. A. Colvin, R. A. Davenport and M. R. Nichols, Arch. Biochem. Biophys. 2016, 597, 1.